|
 
General information:
 Gram-positive spherical bacteria belong to Micrococcaceae family. Gram-positive spherical bacteria belong to Micrococcaceae family.
 Classified into two major groups: aureus and non-aureus. S. aureus is one of the major causes of community-acquired and hospital-acquired infection. Of the non-aureus species, S. epidermis is the most clinically significant. Classified into two major groups: aureus and non-aureus. S. aureus is one of the major causes of community-acquired and hospital-acquired infection. Of the non-aureus species, S. epidermis is the most clinically significant.
Characteristics:
 Primarily an extracellular pathogen. Primarily an extracellular pathogen.
 Adherence is mediated by surface protein adhesins called MSCRAMMs (microbial surface components recognizing adhesive matrix molecules). Adherence is mediated by surface protein adhesins called MSCRAMMs (microbial surface components recognizing adhesive matrix molecules).
 One feature contributes to the virulence of S. epidermidis is the ability to adhere to plasitic and form a biofilm. One feature contributes to the virulence of S. epidermidis is the ability to adhere to plasitic and form a biofilm.
Disease:
 S. aureus: a wide variety of diseases, ranging form superficial abscesses and wound infections to deep and systemic infections such as osteomyelitis, endocarditis and septicaemia. S. aureus: a wide variety of diseases, ranging form superficial abscesses and wound infections to deep and systemic infections such as osteomyelitis, endocarditis and septicaemia.
 Toxic-shock syndrome, staphylococcal scarlet fever, scalded skin syndrome. Toxic-shock syndrome, staphylococcal scarlet fever, scalded skin syndrome.
 S. epidermidis: catheter-associated infections, biofilms on plastic implants, endocarditis. S. epidermidis: catheter-associated infections, biofilms on plastic implants, endocarditis.
Selected genomes: ⇒ comparative pathogenomics ⇐
 S. aureus RF122, 2742531 bp, NC_007622 S. aureus RF122, 2742531 bp, NC_007622
 S. aureus str. O46, 2791410 bp, CP025395 S. aureus str. O46, 2791410 bp, CP025395
 S. aureus str. pt251, 2818393 bp, CP049443 S. aureus str. pt251, 2818393 bp, CP049443
 S. aureus str. UMCG579, 2741379 bp, CP091066 S. aureus str. UMCG579, 2741379 bp, CP091066
 S. aureus subsp. aureus COL, 2809422 bp, NC_002951 S. aureus subsp. aureus COL, 2809422 bp, NC_002951
 S. aureus subsp. aureus JH1, 2906507 bp, NC_009632 S. aureus subsp. aureus JH1, 2906507 bp, NC_009632
 S. aureus subsp. aureus JH9, 2906700 bp, NC_009487 S. aureus subsp. aureus JH9, 2906700 bp, NC_009487
 S. aureus subsp. aureus MRSA252, 2902619 bp, NC_002952 S. aureus subsp. aureus MRSA252, 2902619 bp, NC_002952
 S. aureus subsp. aureus MSSA476, 2799802 bp, NC_002953 S. aureus subsp. aureus MSSA476, 2799802 bp, NC_002953
 S. aureus subsp. aureus Mu3, 2880168 bp, NC_009782 S. aureus subsp. aureus Mu3, 2880168 bp, NC_009782
 S. aureus subsp. aureus Mu50, 2878529 bp, NC_002758 S. aureus subsp. aureus Mu50, 2878529 bp, NC_002758
 S. aureus subsp. aureus MW2, 2820462 bp, NC_003923 S. aureus subsp. aureus MW2, 2820462 bp, NC_003923
 S. aureus subsp. aureus N315, 2814816 bp, NC_002745 S. aureus subsp. aureus N315, 2814816 bp, NC_002745
 S. aureus subsp. aureus NCTC 8325, 2821361 bp, NC_007795 S. aureus subsp. aureus NCTC 8325, 2821361 bp, NC_007795
 S. aureus subsp. aureus str. Newman, 2878897 bp, NC_009641 S. aureus subsp. aureus str. Newman, 2878897 bp, NC_009641
 S. aureus subsp. aureus USA300_FPR3757, 2872769 bp, NC_007793 S. aureus subsp. aureus USA300_FPR3757, 2872769 bp, NC_007793
 S. aureus subsp. aureus USA300_TCH1516, 2872915 bp, NC_010079 S. aureus subsp. aureus USA300_TCH1516, 2872915 bp, NC_010079
 S. epidermidis ATCC 12228, 2499279 bp, NC_004461 S. epidermidis ATCC 12228, 2499279 bp, NC_004461
 S. epidermidis RP62A, 2616530 bp, NC_002976 S. epidermidis RP62A, 2616530 bp, NC_002976
 S. haemolyticus JCSC1435, 2685015 bp, NC_007168 S. haemolyticus JCSC1435, 2685015 bp, NC_007168
 S. saprophyticus subsp. saprophyticus ATCC 15305, 2516575 bp, NC_007350 S. saprophyticus subsp. saprophyticus ATCC 15305, 2516575 bp, NC_007350
VF-related plasmids:
 S. aureus str. TY4 pETB, 38211 bp, NC_003265 S. aureus str. TY4 pETB, 38211 bp, NC_003265
Genome-related publications:
 Kuroda M, et al., 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus Lancet 357(9264):1225-1240. Kuroda M, et al., 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus Lancet 357(9264):1225-1240.
 Yamaguchi T, et al., 2001. Complete nucleotide sequence of a Staphylococcus aureus exfoliative toxin B plasmid and identification of a novel ADP-ribosyltransferase, EDIN-C. Infect Immun 69(12):7760-71. Yamaguchi T, et al., 2001. Complete nucleotide sequence of a Staphylococcus aureus exfoliative toxin B plasmid and identification of a novel ADP-ribosyltransferase, EDIN-C. Infect Immun 69(12):7760-71.
 Baba T, et al., 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359(9320):1819-1827. Baba T, et al., 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359(9320):1819-1827.
 Zhang YQ, et al., 2003. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228) Mol. Microbiol. 49(6):1577-1593. Zhang YQ, et al., 2003. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228) Mol. Microbiol. 49(6):1577-1593.
 Holden MT, et al., 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. USA. 101(26):9786-9791. Holden MT, et al., 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. USA. 101(26):9786-9791.
 Gill SR, et al., 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol 187(7):2426-2438. Gill SR, et al., 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol 187(7):2426-2438.
 Kuroda M, et al., 2005. Whole genome sequence of Staphylococcus saprophyticus reveals the pathogenesis of uncomplicated urinary tract infection. Proc Natl Acad Sci USA 102(37):13272-13277. Kuroda M, et al., 2005. Whole genome sequence of Staphylococcus saprophyticus reveals the pathogenesis of uncomplicated urinary tract infection. Proc Natl Acad Sci USA 102(37):13272-13277.
 Takeuchi F, et al., 2005. Whole-genome sequencing of staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J Bacteriol 187(21):7292-7308. Takeuchi F, et al., 2005. Whole-genome sequencing of staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J Bacteriol 187(21):7292-7308.
 Diep BA, et al., 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367(9512):731-739. Diep BA, et al., 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367(9512):731-739.
 Baba T, et al., 2008. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J Bacteriol 190(1):300-310. Baba T, et al., 2008. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J Bacteriol 190(1):300-310.
 Neoh HM, et al., 2008. Mutated response regulator graR is responsible for phenotypic conversion of Staphylococcus aureus from heterogeneous vancomycin-intermediate resistance to vancomycin-intermediate resistance. Antimicrob Agents Chemother. 52(1):45-53. Neoh HM, et al., 2008. Mutated response regulator graR is responsible for phenotypic conversion of Staphylococcus aureus from heterogeneous vancomycin-intermediate resistance to vancomycin-intermediate resistance. Antimicrob Agents Chemother. 52(1):45-53.
 Herron-Olson L, et al., 2007. Molecular Correlates of Host Specialization in Staphylococcus aureus. PLoS ONE 2(10):e1120. Herron-Olson L, et al., 2007. Molecular Correlates of Host Specialization in Staphylococcus aureus. PLoS ONE 2(10):e1120.
 Highlander SK, et al., 2007. Subtle genetic changes enhance virulence of methicillin resistant and sensitive Staphylococcus aureus. BMC Microbiol 7:99. Highlander SK, et al., 2007. Subtle genetic changes enhance virulence of methicillin resistant and sensitive Staphylococcus aureus. BMC Microbiol 7:99.
 Le Maréchal C, et al., 2011. Genome sequences of two Staphylococcus aureus ovine strains that induce severe (strain O11) and mild (strain O46) mastitis. J Bacteriol 193(9):2353-4. Le Maréchal C, et al., 2011. Genome sequences of two Staphylococcus aureus ovine strains that induce severe (strain O11) and mild (strain O46) mastitis. J Bacteriol 193(9):2353-4.
 Sabat AJ, et al., 2022. Case Report: Necrotizing fasciitis caused by Staphylococcus aureus positive for a new sequence variant of exfoliative toxin E. Front Genet 13:964358. Sabat AJ, et al., 2022. Case Report: Necrotizing fasciitis caused by Staphylococcus aureus positive for a new sequence variant of exfoliative toxin E. Front Genet 13:964358.
Major virulence factors in Staphylococcus:





  




  
  
  





  




















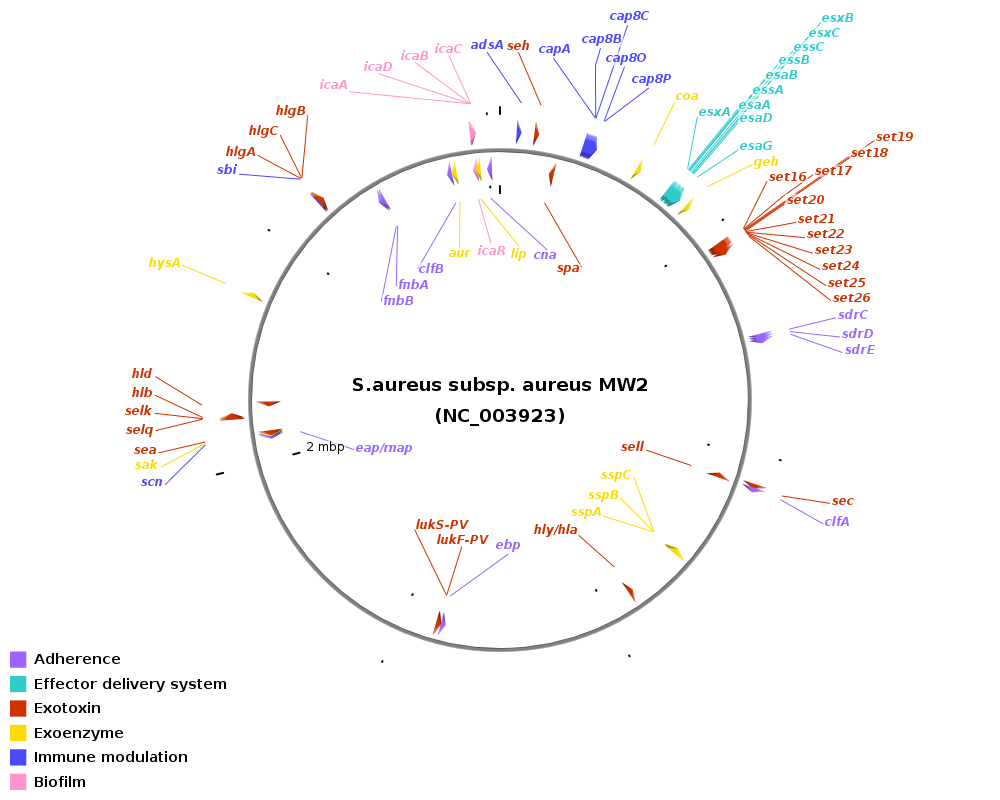
Genomic location of virulence-related genes in Staphylococcus:
 |
Reported anti-virulence compounds to Staphylococcus:






























 |
| Back to Top |
|
