|
 
General information:
 Enterococci are Gram-positive facultative anaerobes inhabiting the gastrointestinal tract, the oral cavity, and the vagina in humans as normal commensals. E. faecalis and E. faecium are the species most commonly associated with colonization of the human gastrointestinal tract. Enterococci are Gram-positive facultative anaerobes inhabiting the gastrointestinal tract, the oral cavity, and the vagina in humans as normal commensals. E. faecalis and E. faecium are the species most commonly associated with colonization of the human gastrointestinal tract.
 Currently, enterococci rank among the top third leading cause of nosocomial infection. The majority of enterococcal infections are caused by either E. faecalis or E. faecium. To a significantly lesser extent, infections are caused by other species of enterocci such as E. durans, E. avium, E. gallinarum, or E. casseliflavus. While E. faecium constitutes the majority of vancomycin and ampicillin resistant isolates of enterococci, E. faecalis accounts for the majority (65-80%) of nosocomial infections caused by enterococci. Currently, enterococci rank among the top third leading cause of nosocomial infection. The majority of enterococcal infections are caused by either E. faecalis or E. faecium. To a significantly lesser extent, infections are caused by other species of enterocci such as E. durans, E. avium, E. gallinarum, or E. casseliflavus. While E. faecium constitutes the majority of vancomycin and ampicillin resistant isolates of enterococci, E. faecalis accounts for the majority (65-80%) of nosocomial infections caused by enterococci.
Characteristics:
 A potential reason for the emergence of E. faecalis as a causative agent of nosocomial infection is the robust nature of this organism. E. faecalis has an intrinsic ability to grow in hypotonic, hypertonic, acidic, or alkaline conditions and to withstand detergents, oxidative stress, and desiccation. A potential reason for the emergence of E. faecalis as a causative agent of nosocomial infection is the robust nature of this organism. E. faecalis has an intrinsic ability to grow in hypotonic, hypertonic, acidic, or alkaline conditions and to withstand detergents, oxidative stress, and desiccation.
 The E. faecalis V583 genome contains 134 predicted surface-exposed proteins, of which 65 contain repeat regions and may have a role in antigenic variation via a slippage mechanism. The E. faecalis V583 genome contains 134 predicted surface-exposed proteins, of which 65 contain repeat regions and may have a role in antigenic variation via a slippage mechanism.
Disease:
 Enterococci: causes of urinary tract infections, bacteremia, peritonitis and infective endocarditis. Enterococci: causes of urinary tract infections, bacteremia, peritonitis and infective endocarditis.
Selected genomes: ⇒ comparative pathogenomics ⇐
 E. faecalis D32, 2987450 bp, NC_018221 E. faecalis D32, 2987450 bp, NC_018221
 E. faecalis OG1RF, 2739625 bp, NC_017316 E. faecalis OG1RF, 2739625 bp, NC_017316
 E. faecalis V583, 3218031 bp, NC_004668 E. faecalis V583, 3218031 bp, NC_004668
 E. faecium Aus0004, 2955294 bp, NC_017022 E. faecium Aus0004, 2955294 bp, NC_017022
 E. faecium Aus0085, 2994661 bp, NC_021994 E. faecium Aus0085, 2994661 bp, NC_021994
 E. faecium DO, 2698137 bp, NC_017960 E. faecium DO, 2698137 bp, NC_017960
 E. faecium NRRL B-2354, 2635572 bp, NC_020207 E. faecium NRRL B-2354, 2635572 bp, NC_020207
VF-related plasmids:
 E. faecalis str. E99 pBEE99, 80600 bp, GU046453 E. faecalis str. E99 pBEE99, 80600 bp, GU046453
 E. faecalis V583 pTEF1, 66320 bp, NC_004669 E. faecalis V583 pTEF1, 66320 bp, NC_004669
 E. faecalis V583 pTEF2, 57660 bp, NC_004671 E. faecalis V583 pTEF2, 57660 bp, NC_004671
Genome-related publications:
 Paulsen I, et al., 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299(5615):2071-2074. Paulsen I, et al., 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299(5615):2071-2074.
 Lam MM, et al., 2012. Comparative analysis of the first complete Enterococcus faecium genome. J Bacteriol 194(9):2334-41. Lam MM, et al., 2012. Comparative analysis of the first complete Enterococcus faecium genome. J Bacteriol 194(9):2334-41.
 Qin X, et al., 2012. Complete genome sequence of Enterococcus faecium strain TX16 and comparative genomic analysis of Enterococcus faecium genomes. BMC Microbiol 12:135. Qin X, et al., 2012. Complete genome sequence of Enterococcus faecium strain TX16 and comparative genomic analysis of Enterococcus faecium genomes. BMC Microbiol 12:135.
 Zischka M, et al., 2012. Complete genome sequence of the porcine isolate Enterococcus faecalis D32. J Bacteriol 194(19):5490-1. Zischka M, et al., 2012. Complete genome sequence of the porcine isolate Enterococcus faecalis D32. J Bacteriol 194(19):5490-1.
 Lam MM, et al., 2013. Comparative analysis of the complete genome of an epidemic hospital sequence type 203 clone of vancomycin-resistant Enterococcus faecium. BMC Genomics 14:595. Lam MM, et al., 2013. Comparative analysis of the complete genome of an epidemic hospital sequence type 203 clone of vancomycin-resistant Enterococcus faecium. BMC Genomics 14:595.
 Kopit LM, et al., 2014. Safety of the surrogate microorganism Enterococcus faecium NRRL B-2354 for use in thermal process validation. Appl Environ Microbiol 80(6):1899-909. Kopit LM, et al., 2014. Safety of the surrogate microorganism Enterococcus faecium NRRL B-2354 for use in thermal process validation. Appl Environ Microbiol 80(6):1899-909.
Major virulence factors in Enterococcus:


 - E. faecium - E. faecium


 - E. faecium - E. faecium


 - E. faecium - E. faecium
 - E. faecium - E. faecium











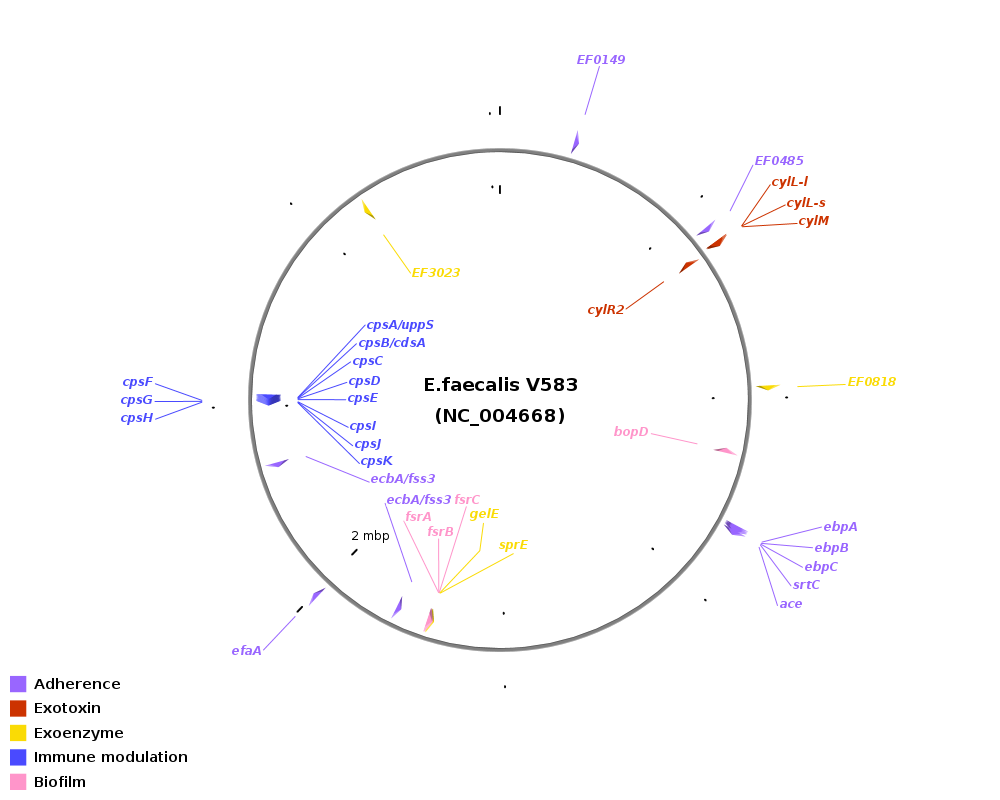
Genomic location of virulence-related genes in Enterococcus:
 |
Reported anti-virulence compounds to Enterococcus:



















 |
| Back to Top |
|
