|
General information:
 A genus of bacteria in the family of Mycobacteriaceae, Gram-positive, the majority of the over 50 species ar non-pathogenic environmental bacteria related closely to the soil bacteria Streptomyces and Actinomyces. A genus of bacteria in the family of Mycobacteriaceae, Gram-positive, the majority of the over 50 species ar non-pathogenic environmental bacteria related closely to the soil bacteria Streptomyces and Actinomyces.
 A few species are highly successful pathogens, including Mycobacterium tuberculosis, M. leprae and M. ulcerans. A few species are highly successful pathogens, including Mycobacterium tuberculosis, M. leprae and M. ulcerans.
Characteristics:
 M. tuberculosis uses the respiratory tract as the principal portal of entry, where it travels deep into the alveoli and is engulfed by alveolar macrophages. M. tuberculosis can persist and even replicate in this extremely hostile intracellular enviroment. M. tuberculosis uses the respiratory tract as the principal portal of entry, where it travels deep into the alveoli and is engulfed by alveolar macrophages. M. tuberculosis can persist and even replicate in this extremely hostile intracellular enviroment.
 Pathogenic mycobacteria are extraordinary adept at establishing long-term infections that can manifest as acute or chonic disease or be clinically asymptomatic with the potential to resurface later. Pathogenic mycobacteria are extraordinary adept at establishing long-term infections that can manifest as acute or chonic disease or be clinically asymptomatic with the potential to resurface later.
 One of the major features of M. tuberculosis is its capacity to block the acidification of the phagosome and consequently to disable or retard phagosome-lysosome fusion in phagocytic cells. Complex lipids from the mycobacterial cell wall are thought to be important for blocking the normal biogenesis of the phagolysosome. One of the major features of M. tuberculosis is its capacity to block the acidification of the phagosome and consequently to disable or retard phagosome-lysosome fusion in phagocytic cells. Complex lipids from the mycobacterial cell wall are thought to be important for blocking the normal biogenesis of the phagolysosome.
Disease:
 M. tuberculosis is the causative agents of tuberculosis. M. tuberculosis is the causative agents of tuberculosis.
 M. leprae: leprosy. M. leprae: leprosy.
 M. ulcerans: Buruli ulcers. M. ulcerans: Buruli ulcers.
Selected genomes: ⇒ comparative pathogenomics ⇐
 M. abscessus ATCC 19977 chromosome 1, 5067172 bp, NC_010397 M. abscessus ATCC 19977 chromosome 1, 5067172 bp, NC_010397
 M. abscessus subsp. bolletii 50594, 5000473 bp, NC_021282 M. abscessus subsp. bolletii 50594, 5000473 bp, NC_021282
 M. abscessus subsp. bolletii str. GO 06, 5068807 bp, NC_018150 M. abscessus subsp. bolletii str. GO 06, 5068807 bp, NC_018150
 M. africanum GM041182, 4389314 bp, NC_015758 M. africanum GM041182, 4389314 bp, NC_015758
 M. avium 104, 5475491 bp, NC_008595 M. avium 104, 5475491 bp, NC_008595
 M. avium subsp. paratuberculosis K-10, 4829781 bp, NC_002944 M. avium subsp. paratuberculosis K-10, 4829781 bp, NC_002944
 M. avium subsp. paratuberculosis MAP4, 4829424 bp, NC_021200 M. avium subsp. paratuberculosis MAP4, 4829424 bp, NC_021200
 M. bovis AF2122/97, 4345492 bp, NC_002945 M. bovis AF2122/97, 4345492 bp, NC_002945
 M. bovis BCG str. Korea 1168P, 4376711 bp, NC_020245 M. bovis BCG str. Korea 1168P, 4376711 bp, NC_020245
 M. bovis BCG str. Mexico, 4350386 bp, NC_016804 M. bovis BCG str. Mexico, 4350386 bp, NC_016804
 M. bovis BCG str. Pasteur 1173P2, 4374522 bp, NC_008769 M. bovis BCG str. Pasteur 1173P2, 4374522 bp, NC_008769
 M. bovis BCG str. Tokyo 172, 4371711 bp, NC_012207 M. bovis BCG str. Tokyo 172, 4371711 bp, NC_012207
 M. canettii CIPT 140010059, 4482059 bp, NC_015848 M. canettii CIPT 140010059, 4482059 bp, NC_015848
 M. canettii CIPT 140060008, 4432426 bp, NC_019950 M. canettii CIPT 140060008, 4432426 bp, NC_019950
 M. canettii CIPT 140070008, 4420197 bp, NC_019965 M. canettii CIPT 140070008, 4420197 bp, NC_019965
 M. canettii CIPT 140070010, 4525948 bp, NC_019951 M. canettii CIPT 140070010, 4525948 bp, NC_019951
 M. canettii CIPT 140070017, 4524466 bp, NC_019952 M. canettii CIPT 140070017, 4524466 bp, NC_019952
 M. gilvum PYR-GCK, 5619607 bp, NC_009338 M. gilvum PYR-GCK, 5619607 bp, NC_009338
 M. gilvum Spyr1, 5547747 bp, NC_014814 M. gilvum Spyr1, 5547747 bp, NC_014814
 M. indicus pranii MTCC 9506, 5589007 bp, NC_018612 M. indicus pranii MTCC 9506, 5589007 bp, NC_018612
 M. intracellulare ATCC 13950, 5402402 bp, NC_016946 M. intracellulare ATCC 13950, 5402402 bp, NC_016946
 M. intracellulare MOTT-02, 5409696 bp, NC_016947 M. intracellulare MOTT-02, 5409696 bp, NC_016947
 M. intracellulare MOTT-64, 5501090 bp, NC_016948 M. intracellulare MOTT-64, 5501090 bp, NC_016948
 M. intracellulare str. MOTT36Y, 5613626 bp, NC_017904 M. intracellulare str. MOTT36Y, 5613626 bp, NC_017904
 M. leprae Br4923, 3268071 bp, NC_011896 M. leprae Br4923, 3268071 bp, NC_011896
 M. leprae TN, 3268203 bp, NC_002677 M. leprae TN, 3268203 bp, NC_002677
 M. liflandii 128FXT, 6208955 bp, NC_020133 M. liflandii 128FXT, 6208955 bp, NC_020133
 M. marinum M, 6636827 bp, NC_010612 M. marinum M, 6636827 bp, NC_010612
 M. smegmatis JS623, 6464916 bp, NC_019966 M. smegmatis JS623, 6464916 bp, NC_019966
 M. smegmatis str. MC2 155, 6988209 bp, NC_008596 M. smegmatis str. MC2 155, 6988209 bp, NC_008596
 M. sp. JDM601, 4643668 bp, NC_015576 M. sp. JDM601, 4643668 bp, NC_015576
 M. sp. JLS, 6048425 bp, NC_009077 M. sp. JLS, 6048425 bp, NC_009077
 M. sp. KMS, 5737227 bp, NC_008705 M. sp. KMS, 5737227 bp, NC_008705
 M. sp. MCS, 5705448 bp, NC_008146 M. sp. MCS, 5705448 bp, NC_008146
 M. tuberculosis 7199-99, 4421197 bp, NC_020089 M. tuberculosis 7199-99, 4421197 bp, NC_020089
 M. tuberculosis CAS/NITR204, 4392876 bp, NC_021193 M. tuberculosis CAS/NITR204, 4392876 bp, NC_021193
 M. tuberculosis CCDC5079, 4398812 bp, NC_017523 M. tuberculosis CCDC5079, 4398812 bp, NC_017523
 M. tuberculosis CCDC5180, 4405981 bp, NC_017522 M. tuberculosis CCDC5180, 4405981 bp, NC_017522
 M. tuberculosis CDC1551, 4403837 bp, NC_002755 M. tuberculosis CDC1551, 4403837 bp, NC_002755
 M. tuberculosis CTRI-2, 4398525 bp, NC_017524 M. tuberculosis CTRI-2, 4398525 bp, NC_017524
 M. tuberculosis F11, 4424435 bp, NC_009565 M. tuberculosis F11, 4424435 bp, NC_009565
 M. tuberculosis H37Ra, 4419977 bp, NC_009525 M. tuberculosis H37Ra, 4419977 bp, NC_009525
 M. tuberculosis H37Rv, 4411532 bp, NC_000962 M. tuberculosis H37Rv, 4411532 bp, NC_000962
 M. tuberculosis KZN 1435, 4398250 bp, NC_012943 M. tuberculosis KZN 1435, 4398250 bp, NC_012943
 M. tuberculosis KZN 4207, 4394985 bp, NC_016768 M. tuberculosis KZN 4207, 4394985 bp, NC_016768
 M. tuberculosis KZN 605, 4399120 bp, NC_018078 M. tuberculosis KZN 605, 4399120 bp, NC_018078
 M. tuberculosis RGTB327, 4380119 bp, NC_017026 M. tuberculosis RGTB327, 4380119 bp, NC_017026
 M. tuberculosis RGTB423, 4406587 bp, NC_017528 M. tuberculosis RGTB423, 4406587 bp, NC_017528
 M. tuberculosis str. Beijing/NITR203, 4411128 bp, NC_021054 M. tuberculosis str. Beijing/NITR203, 4411128 bp, NC_021054
 M. tuberculosis str. Erdman (ATCC 35801), 4392353 bp, NC_020559 M. tuberculosis str. Erdman (ATCC 35801), 4392353 bp, NC_020559
 M. tuberculosis str. Haarlem/NITR202, 4404786 bp, NC_021192 M. tuberculosis str. Haarlem/NITR202, 4404786 bp, NC_021192
 M. tuberculosis UT205, 4418088 bp, NC_016934 M. tuberculosis UT205, 4418088 bp, NC_016934
 M. ulcerans Agy99, 5631606 bp, NC_008611 M. ulcerans Agy99, 5631606 bp, NC_008611
 M. vanbaalenii PYR-1, 6491865 bp, NC_008726 M. vanbaalenii PYR-1, 6491865 bp, NC_008726
 M. yongonense 05-1390, 5521023 bp, NC_021715 M. yongonense 05-1390, 5521023 bp, NC_021715
Plasmids:
 M. gilvum Spyr1 pMSPYR101, 211864 bp, NC_014811 M. gilvum Spyr1 pMSPYR101, 211864 bp, NC_014811
 M. liflandii 128FXT pMUM002, 190588 bp, NC_011355 M. liflandii 128FXT pMUM002, 190588 bp, NC_011355
 M. ulcerans Agy99 pMUM001, 174155 bp, NC_005916 M. ulcerans Agy99 pMUM001, 174155 bp, NC_005916
Related publications:
 Cole ST, et al., 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393(6685):537-544. Cole ST, et al., 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393(6685):537-544.
 Cole ST, et al., 2001. Massive gene decay in the leprosy bacillus. Nature 409(6823):1007-1011. Cole ST, et al., 2001. Massive gene decay in the leprosy bacillus. Nature 409(6823):1007-1011.
 Fleischmann RD, et al., 2002. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 184(19):5479-5490. Fleischmann RD, et al., 2002. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 184(19):5479-5490.
 Garnier T, et al., 2003. The complete genome sequence of Mycobacterium bovis Proc. Natl. Acad. Sci. USA. 100(13):7877-7882. Garnier T, et al., 2003. The complete genome sequence of Mycobacterium bovis Proc. Natl. Acad. Sci. USA. 100(13):7877-7882.
 Stinear TP, et al., 2004. Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proc Natl Acad Sci USA 101(5):1345-1349. Stinear TP, et al., 2004. Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proc Natl Acad Sci USA 101(5):1345-1349.
 Li L, et al., 2005. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc Natl Acad Sci USA 102(35):12344-12349. Li L, et al., 2005. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc Natl Acad Sci USA 102(35):12344-12349.
 Stinear TP, et al., 2007. Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res 17(2):192-200. Stinear TP, et al., 2007. Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res 17(2):192-200.
 Brosch R, et al., 2007. Genome plasticity of BCG and impact on vaccine efficacy. Proc Natl Acad Sci USA 104(13):5596-5601. Brosch R, et al., 2007. Genome plasticity of BCG and impact on vaccine efficacy. Proc Natl Acad Sci USA 104(13):5596-5601.
 Stinear TP, et al., 2008. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res 18(5):729-741. Stinear TP, et al., 2008. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res 18(5):729-741.
 Zheng H, et al., 2008. Genetic basis of virulence attenuation revealed by comparative genomic analysis of Mycobacterium tuberculosis strain H37Ra versus H37Rv. PLoS ONE 3(6):e2375. Zheng H, et al., 2008. Genetic basis of virulence attenuation revealed by comparative genomic analysis of Mycobacterium tuberculosis strain H37Ra versus H37Rv. PLoS ONE 3(6):e2375.
 Pidot SJ, et al., 2008. Deciphering the genetic basis for polyketide variation among mycobacteria producing mycolactones. BMC Genomics. 9:462. Pidot SJ, et al., 2008. Deciphering the genetic basis for polyketide variation among mycobacteria producing mycolactones. BMC Genomics. 9:462.
 Seki M, et al., 2009. Whole genome sequence analysis of Mycobacterium bovis bacillus Calmette-Guérin (BCG) Tokyo 172: a comparative study of BCG vaccine substrains. Vaccine 27(11):1710-1716. Seki M, et al., 2009. Whole genome sequence analysis of Mycobacterium bovis bacillus Calmette-Guérin (BCG) Tokyo 172: a comparative study of BCG vaccine substrains. Vaccine 27(11):1710-1716.
 Ripoll F, et al., 2009. Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS One 4(6):e5660. Ripoll F, et al., 2009. Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS One 4(6):e5660.
 Monot M, et al., 2009. Comparative genomic and phylogeographic analysis of Mycobacterium leprae. Nat Genet 41(12):1282-1289. Monot M, et al., 2009. Comparative genomic and phylogeographic analysis of Mycobacterium leprae. Nat Genet 41(12):1282-1289.
 Zhang ZY, et al., 2011. Complete genome sequence of a novel clinical isolate, the nontuberculous Mycobacterium strain JDM601. J Bacteriol 193(16):4300-4301. Zhang ZY, et al., 2011. Complete genome sequence of a novel clinical isolate, the nontuberculous Mycobacterium strain JDM601. J Bacteriol 193(16):4300-4301.
 Zhang Y, et al., 2011. Complete genome sequences of Mycobacterium tuberculosis strains CCDC5079 and CCDC5080, which belong to the Beijing family. J Bacteriol 193(19):5591-2. Zhang Y, et al., 2011. Complete genome sequences of Mycobacterium tuberculosis strains CCDC5079 and CCDC5080, which belong to the Beijing family. J Bacteriol 193(19):5591-2.
 Orduna P, et al., 2011. Genomic and proteomic analyses of Mycobacterium bovis BCG Mexico 1931 reveal a diverse immunogenic repertoire against tuberculosis infection. BMC Genomics 12:493. Orduna P, et al., 2011. Genomic and proteomic analyses of Mycobacterium bovis BCG Mexico 1931 reveal a diverse immunogenic repertoire against tuberculosis infection. BMC Genomics 12:493.
 Kallimanis A, et al., 2011. Complete genome sequence of Mycobacterium sp. strain (Spyr1) and reclassification to Mycobacterium gilvum Spyr1. Stand Genomic Sci 5(1):144-153. Kallimanis A, et al., 2011. Complete genome sequence of Mycobacterium sp. strain (Spyr1) and reclassification to Mycobacterium gilvum Spyr1. Stand Genomic Sci 5(1):144-153.
 Isaza JP, et al., 2012. Whole genome shotgun sequencing of one Colombian clinical isolate of Mycobacterium tuberculosis reveals DosR regulon gene deletions. FEMS Microbiol Lett 330(2):113-20. Isaza JP, et al., 2012. Whole genome shotgun sequencing of one Colombian clinical isolate of Mycobacterium tuberculosis reveals DosR regulon gene deletions. FEMS Microbiol Lett 330(2):113-20.
 Kim BJ, et al., 2012. Complete genome sequence of Mycobacterium intracellulare strain ATCC 13950(T). J Bacteriol 194(10):2750-2750. Kim BJ, et al., 2012. Complete genome sequence of Mycobacterium intracellulare strain ATCC 13950(T). J Bacteriol 194(10):2750-2750.
 Miyoshi-Akiyama T, et al., 2012. Complete annotated genome sequence of Mycobacterium tuberculosis Erdman. J Bacteriol 194(10):2770. Miyoshi-Akiyama T, et al., 2012. Complete annotated genome sequence of Mycobacterium tuberculosis Erdman. J Bacteriol 194(10):2770.
 Kim BJ, et al., 2012. Complete genome sequence of Mycobacterium intracellulare clinical strain MOTT-02. J Bacteriol 194(10):2771-2771. Kim BJ, et al., 2012. Complete genome sequence of Mycobacterium intracellulare clinical strain MOTT-02. J Bacteriol 194(10):2771-2771.
 Kim BJ, et al., 2012. Complete genome sequence of Mycobacterium intracellulare clinical strain MOTT-64, belonging to the INT1 genotype. J Bacteriol 194(12):3268-3268. Kim BJ, et al., 2012. Complete genome sequence of Mycobacterium intracellulare clinical strain MOTT-64, belonging to the INT1 genotype. J Bacteriol 194(12):3268-3268.
 Kim BJ, et al., 2012. Complete genome sequence of Mycobacterium intracellulare clinical strain MOTT-36Y, belonging to the INT5 genotype. J Bacteriol 194(15):4141-4142. Kim BJ, et al., 2012. Complete genome sequence of Mycobacterium intracellulare clinical strain MOTT-36Y, belonging to the INT5 genotype. J Bacteriol 194(15):4141-4142.
 Madhavilatha GK, et al., 2012. Whole-genome sequences of two clinical isolates of Mycobacterium tuberculosis from Kerala, South India. J Bacteriol 194(16):4430. Madhavilatha GK, et al., 2012. Whole-genome sequences of two clinical isolates of Mycobacterium tuberculosis from Kerala, South India. J Bacteriol 194(16):4430.
 Raiol T, et al., 2012. Complete genome sequence of Mycobacterium massiliense. J Bacteriol 194(19):5455. Raiol T, et al., 2012. Complete genome sequence of Mycobacterium massiliense. J Bacteriol 194(19):5455.
 Saini V, et al., 2012. Massive gene acquisitions in Mycobacterium indicus pranii provide a perspective on mycobacterial evolution. Nucleic Acids Res 40(21):10832-10850. Saini V, et al., 2012. Massive gene acquisitions in Mycobacterium indicus pranii provide a perspective on mycobacterial evolution. Nucleic Acids Res 40(21):10832-10850.
 Tobias NJ, et al., 2013. Complete genome sequence of the frog pathogen Mycobacterium ulcerans ecovar Liflandii. J Bacteriol 195(3):556-64. Tobias NJ, et al., 2013. Complete genome sequence of the frog pathogen Mycobacterium ulcerans ecovar Liflandii. J Bacteriol 195(3):556-64.
 Supply P, et al., 2013. Genomic analysis of smooth tubercle bacilli provides insights into ancestry and pathoadaptation of Mycobacterium tuberculosis. Nat Genet 45(2):172-9. Supply P, et al., 2013. Genomic analysis of smooth tubercle bacilli provides insights into ancestry and pathoadaptation of Mycobacterium tuberculosis. Nat Genet 45(2):172-9.
 Roetzer A, et al., 2013. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: a longitudinal molecular epidemiological study. PLoS Med 10(2):e1001387. Roetzer A, et al., 2013. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: a longitudinal molecular epidemiological study. PLoS Med 10(2):e1001387.
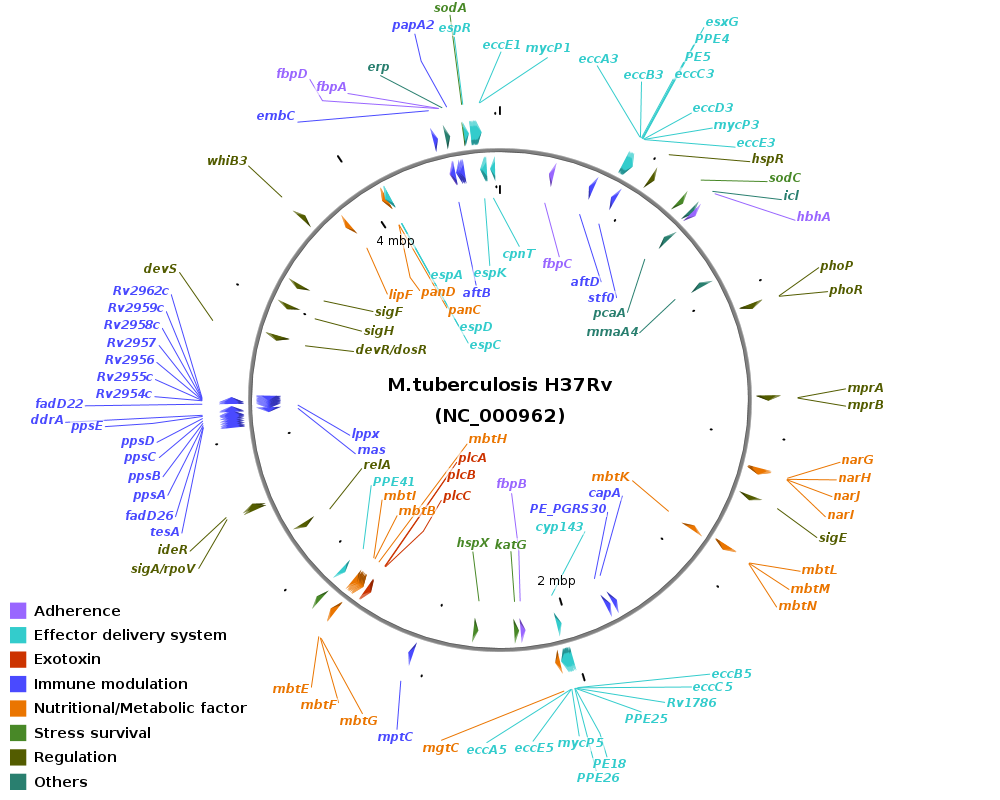
Major virulence factors in Mycobacterium:














































Genomic location of virulence-related genes in Mycobacterium:
 |
 |
| Back to Top |
|
